This video is a summary of my research into how diet might modify the inflammatory response in Alzheimer’s disease and how this could accelerate the disease.
Author Archives: jackrrivers
Invited speaker at the House of Lords
A number FBMH academics and students played a central role in a high profile event which took place recently at the House of Lords. The event, arranged by the Division of Development and Alumni Relations (DDAR), brought together over 200 individuals in an effort to engage with a wider pool of alumni and friends and highlight a snapshot of exciting research and activity.
I spoke about the research he carries out with collaborators, Dr Catherine Lawrence and Dr David Brough, investigating the role of the inflammasome in Alzheimer’s disease and age related cognitive decline. Details of the event can be found here.
University of Manchester Stories Event. Usain Bolt Shaves His Legs
The organisers invited excellent speakers to tell their stories to inspire the next generation of thinkers. Speakers included Dame Prof. Nancy Rothwell and Chancellor Lemn Sissay. The title of my talk was: Usain Bolt shaves his legs.
Statistics example datasets
AAIC 2018
USE OF COMMON PAIN RELIEVING DRUGS CORRELATES WITH ALTERED PROGRESSION OF ALZHEIMER’S DISEASE AND MILD COGNITIVE IMPAIRMENT
Jack Rivers-Auty , Alison E. Mather , Ruth Peters , Catherine B. Lawrence , David Brough,
Background: Our understanding of the pathophysiological mechanisms of Alzheimer’s disease (AD) remains relatively unclear; however, the role of neuroinflammation as a key etiological feature is now widely accepted due to the consensus of epidemiological, neuroimaging, preclinical and genetic evidence. Consequently, non-steroidal anti-inflammatories (NSAIDs) have been investigated in epidemiological and clinical studies as potential disease modifying agents. Previous epidemiological studies focused on incidence of AD and did not thoroughly parse the effects at the individual drug level. The therapeutic potential of modifying incidence has a number of limitations, and we now know that each NSAID subtype has a unique profile of physiological impacts corresponding to different therapeutic profiles for AD. Therefore, we utilized the AD Neuroimaging Initiative (ADNI) dataset to investigate how the use of common NSAIDs and paracetamol alter cognitive decline in subjects with mild cognitive impairment (MCI) or AD.
Methods: Negative binomial generalized linear mixed modelling was utilized to model the cognitive decline of 1619 individuals from the ADNI dataset. Both the mini-mental state examination (MMSE) and AD assessment scale (ADAS) were investigated. Explanatory variables were included or excluded from the model in a stepwise fashion with Chi-square log-likelihood and Akaike information criteria used as selection criteria. Explanatory variables investigated were APOE4, age, diagnosis (control, MCI or AD), gender, education level, vascular pathology, diabetes and drug use (naproxen, celecoxib, diclofenac, aspirin, ibuprofen or paracetamol).
Results: The NSAIDs, aspirin, ibuprofen, naproxen and celecoxib did not significantly alter cognitive decline. However, diclofenac use correlated with slower cognitive decline (ADAS χ2= 4.0, p=0.0455, MMSE χ2= 4.8, p=0.029). Paracetamol use correlated with accelerated decline (ADAS χ2= ¼6.6, p=0.010, MMSE χ2= 8.4, p=0.004). The APOE4 allele correlated with accelerated cognitive deterioration (ADAS χ2= 316.0, p<0.0001, MMSE χ2= 191.0, p<0.0001).
Conclusions: This study thoroughly investigated the effects of common NSAIDs and paracetamol on cognitive decline in MCI and AD subjects. Most common NSAIDs did not alter cognitive decline. However, diclofenac use correlated with slowed cognitive deterioration, providing exciting evidence for a potential disease modifying therapeutic. Conversely, paracetamol use correlated with accelerated decline; which, if confirmed to be causative, would have massive ramifications for the recommended use of this prolific drug.
Exploration of RNAseq and Epidemiological datasets
Utilizing R statistical software and Shiny R package I’ve developed web apps for the exploration of RNAseq and epidemiological datasets.
Not all NSAIDs are the same: Differing effects of pain-relieving drugs on cognitive decline in the Alzheimer’s Disease Neuroimaging Initiative dataset.
https://braininflammationgroup-universityofmanchester.shinyapps.io/Rivers-Auty-ADNI/
Citation:
Rivers-Auty J, Mather AE, Peters R, Lawrence CB, Brough D (2018) Use of common pain relieving drugs correlates with altered progression of Alzheimer’s disease and mild cognitive impairment. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association 14 (7):P1337-P1338
Small, Thin Graphene Oxide is Anti-inflammatory Activating NRF2 via Metabolic Reprogramming. LPS iBMDMs with and without graphene oxide.
https://braininflammationgroup-universityofmanchester.shinyapps.io/GrapheneOxide/
Citation:
Hoyle C, Rivers-Auty J, Lemarchand E, Vranic S, Wang E, Buggio M, Rothwell N, Allan S, Kostarelos K, Brough D (2018) Small, Thin Graphene Oxide Is Anti-Inflammatory Activating Nuclear Factor Erythroid 2-Related Factor 2 (NRF2) Via Metabolic Reprogramming. ACS Nano. DOI: 10.1021/acsnano.8b03642
Haley et al. WT microglia vs NLRP3-/- microglia (not cage mates) https://braininflammationgroup-universityofmanchester.shinyapps.io/NLRP3KOmicroglia/
Dewhurst et al. Liver homogenate and hepatocytes with and without gene deletion https://braininflammationgroup-universityofmanchester.shinyapps.io/Dewhurst-et-al-Correlation/
Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression
https://braininflammationgroup-universityofmanchester.shinyapps.io/Shaw-et-al-TIM-Macs/
Citation:
Shaw TN, Houston SA, Wemyss K, Bridgeman HM, Barbera TA, Zangerle-Murray T, Strangward P, Ridley AJL, Wang P, Tamoutounour S, Allen JE, Konkel JE, Grainger JR (2018) Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Med 215 (6):1507-1518. doi:10.1084/jem.20180019
Zinc deficiency and Alzheimer’s disease.
- Hippocampus homogenate – effects of APP/PS1 genotype and zinc deficiency.
- Means+SEM https://braininflammationgroup-universityofmanchester.shinyapps.io/zinc_deficiency_in_alzheimers_disease_whole_hippocampus/
- Correlations https://braininflammationgroup-universityofmanchester.shinyapps.io/Correlation-Zinc-Deficiency-Alzheimers/
- Plaque vs non-plaque tissue – effects of APP/PS1 genotype and zinc deficiency. https://braininflammationgroup-universityofmanchester.shinyapps.io/Plaque-NLRP3-Alzheimer-Zinc/
Citation:
Rivers-Auty J, White C, Beattie J, Brough D, and Lawrence C. 2017. [Poster Presentation] Zinc deficiency accelerates Alzheimer’s phenotype in the APP/PS1 mouse model. Alzheimer’s Research UK Conference. Aberdeen, UK.
Generalized mixed modelling
Sometimes you know your data can’t follow a normal distribution even after transformation – such as count data or yes/no data. If this is the case, then generalized mixed modelling is the way to go. Basically it’s very similar to general linear modelling which assumes normality of the population distribution, but instead it assumes a different underlying distribution (and it models using maximum likelihood, not minimizing the residuals). For this analysis we have count data (the number of neutrophils in the brain) and a random effect of siblings. Siblings is treated as a random effect because with experiments involving large numbers that come from one parent pair (like fish or fly data), we tend to think that the siblings aren’t independent and perhaps the parents are effecting the outcome. Therefore, we model this lack of independence by treating siblings as a random effect. In fish data we call a bunch of siblings a “clutch”.
To do the analysis, first select the best model. For count data Poisson is the obvious choice, but there are several potential link functions (similar to transformations). So we must select the optimal link function for the Poisson model. However, Poisson has the assumption that the mean is equal to the variance. This is true if each count is completely independent, like counting cars on a quiet country road. However, in biology this is often not the case, in this example having one neutrophil increases your chances of having two neutrophils because an inflammatory response is happening. This is more like a busy road where one car driving by increases the chances of a second car driving by (is it rush hour?), the counts aren’t independent. In this case a negative binomial is a good family to model the data. It has two parameterization methods (the method by which it predicts the lack of independence of the counts). So now you have to model the Poisson models with the three link functions and the negative binomial model with the two parameterization methods and then see which is best. For this we use the Akaike Information Criteria (AIC). With AIC the lower the better, so we will select the model with the lowest AIC.
#Packages
require("lme4")
require("ggplot2")
require("glmmADMB")
#Loading data
example.count<- read.table(url("https://jackauty.com/wp-content/uploads/2018/10/count.txt"), header=T)
str(example.count)
#Generalized mixed modelling
try(glmm.a<-glmmadmb(Neutrophils~Factor1+ (1|Clutch), data=example.count,family="nbinom"))
try(glmm.b<-glmmadmb(Neutrophils~Factor1+ (1|Clutch), data=example.count,family="nbinom1"))
try(glmm.c<-glmer(Neutrophils~Factor1+ (1|Clutch), data=example.count, family=poisson(link="log")))
try(glmm.d<-glmer(Neutrophils~Factor1+ (1|Clutch), data=example.count, family=poisson(link="identity")))
try(glmm.e<-glmer(Neutrophils~Factor1+ (1|Clutch), data=example.count,family=poisson(link="sqrt")))
AIC(glmm.a,glmm.b,glmm.c,glmm.d,glmm.e)
df AIC
glmm.a 4 270.5040
glmm.b 4 273.9180
glmm.c 3 446.7101
glmm.d 3 475.2077
glmm.e 3 461.6097
From this we can see that the negative binomial model is much better and the “nbinom” parameterization is preferred. Now we can continue with our analysis very much like a normal multi-level linear model.
#Build model with and without our independent variable (Factor1) glmm1<-glmmadmb(Neutrophils~1 + (1|Clutch), data=example.count,family="nbinom") glmm2<-update(glmm1, .~. +Factor1) anova(glmm1,glmm2) Analysis of Deviance Table Model 1: Neutrophils ~ 1 Model 2: Neutrophils ~ +Factor1 NoPar LogLik Df Deviance Pr(>Chi) 1 3 -134.27 2 4 -131.25 1 6.028 0.01408 * ---
Our factor significantly improves the model! From this we might say in our publication – “There was a significant effect of factor on neutrophil count (χ2(1)=6.028, p=0.014).”. But first! We have to check the assumptions, this is a little bit different to multi-level linear modelling.
- Is the data approximately distributed appropriately (negative binomial)?
- Are there outlying data points?
- Are the residuals centered around zero for each factor?
- Is there zero inflation (more zeros than expected, this is common in biological data)?
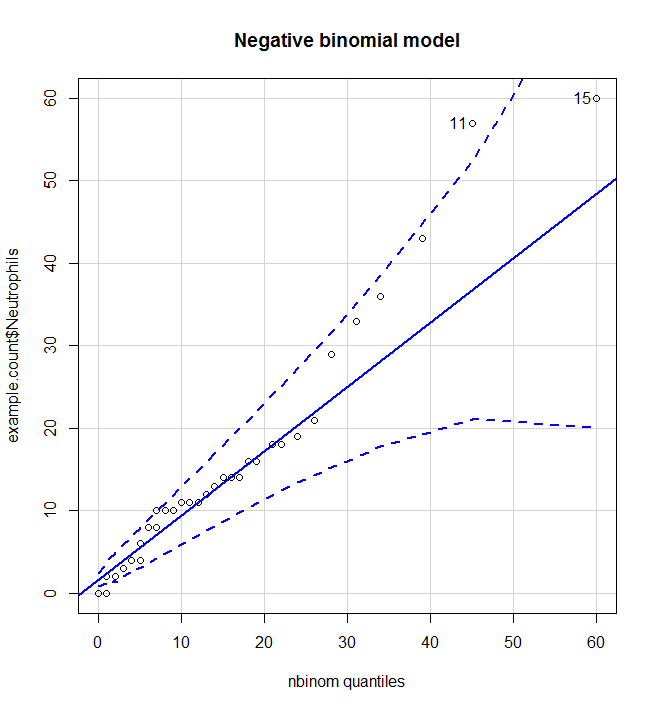
Is the data approximately distributed appropriately (negative binomial)?
From this we can see that the negative binomial model is much better than the Poisson model and most data points lie between the 95% CI of the expected distribution.
#Is the data approximately distributed appropriately (negative binomial)? poisson <- fitdistr(example.count$Neutrophils, "Poisson") qqp(example.count$Neutrophils, "pois", lambda=poisson$estimate, main="Poisson model")nbinom<-fitdistr(example.count$Neutrophils, "negative binomial") qqp(example.count$Neutrophils, "nbinom", size=nbinom$estimate[[1]], mu=nbinom$estimate[[2]], main="Negative binomial model")
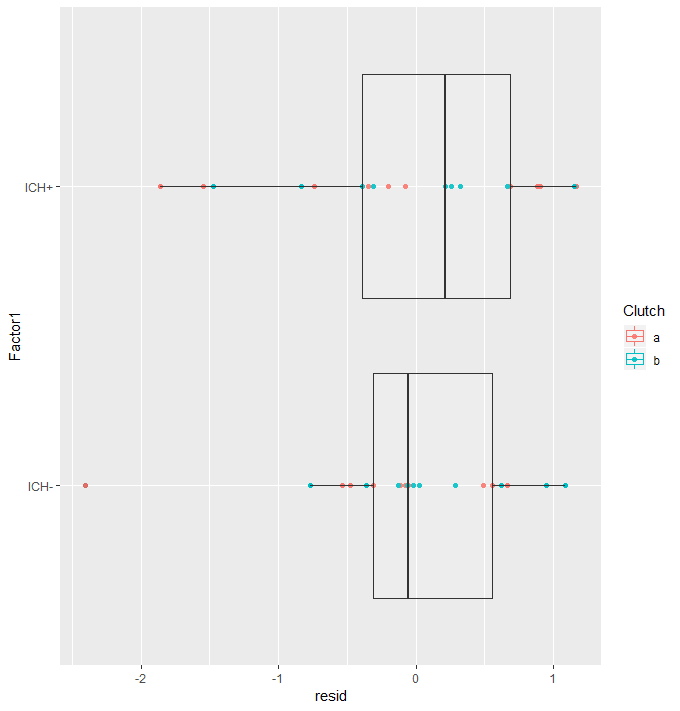
Are there outlying data points? AND Are the residuals centered around zero for each factor?
Plotting the residuals against Factor we can see if the model fitted sensible coefficients and if there are outlying data points. Here we are looking to see if the residuals cluster around zero and that there are no extreme residuals.
Here we can see that the residuals around both factor levels are centered around zero. The random effect of “clutch” seems to model well, with no clustering. There is one residual with an absolute value greater than two. This is slightly concerning with a data set this small. It could be worth checking your lab notes for anything peculiar. I don’t tend to remove data points without a practical reason, for example I might read in my lab notes that this zebra fish looked a lot like a mouse, then I would remove it.
#Are there outlying data points? AND Are the residuals centered around zero for each factor? augDat <- data.frame(example.count,resid=residuals(glmm2,type="pearson"), fitted=fitted(glmm2)) ggplot(augDat,aes(x=Factor1,y=resid,col=Clutch))+geom_point()+geom_boxplot(aes(group=Factor1),alpha = 0.1)+coord_flip()
Is there zero inflation (more zeros than expected, this is common in biological data)?
Here we run the same model but with zeroInflation=T, then we check the AIC and find that the zero-inflation model is better! This sucks because now we have to run the whole thing again with zeroInflation=T. On the re-run, the p-value “improved” and outliers were gone, showing that they were due to zero inflation (and not my zebra fish being a mouse)! So I guess it all works out in the end.
#Is there zero inflation
try(glmm.zero<-glmmadmb(Neutrophils~Factor1+ (1|Clutch), data=example.count,family="nbinom", zeroInflation = T))
AIC(glmm.zero,glmm2)
df AIC
glmm.zero 5 268.298
glmm2 4 270.504
#Quick rerun
glmm1.zero<-glmmadmb(round(Neutrophils)~1 + (1|Clutch), data=example.count,family="nbinom", zeroInflation = T)
glmm2.zero<-update(glmm1.zero, .~. +Factor1)
anova(glmm1.zero,glmm2.zero)
Analysis of Deviance Table
Model 1: round(Neutrophils) ~ 1
Model 2: round(Neutrophils) ~ +Factor1
NoPar LogLik Df Deviance Pr(>Chi)
4 -134.14
5 -129.15 1 9.976 0.001586 **
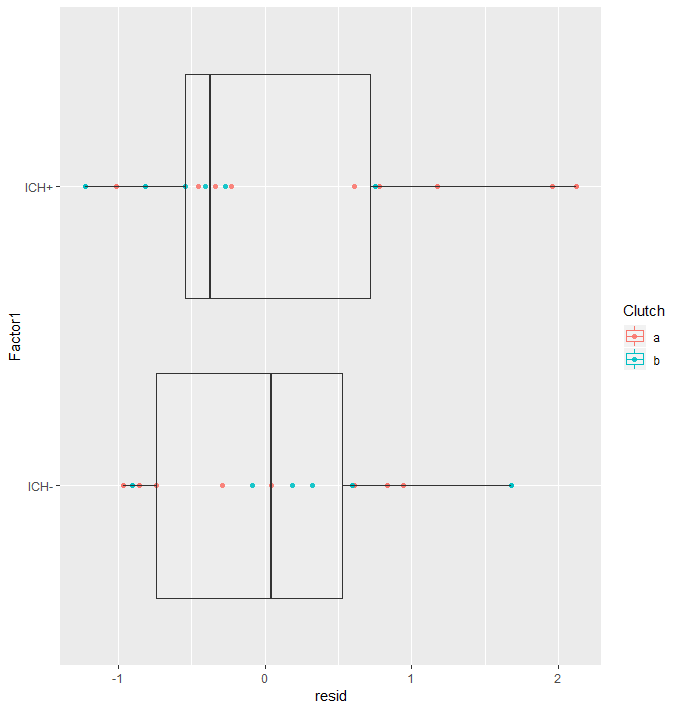
#Quick assumption check
#Are there outlying data points? AND Are the residuals centered around zero for each factor?
augDat <- data.frame(data,resid=residuals(glmm2,type="pearson"),
fitted=fitted(glmm2))
ggplot(augDat,aes(x=Factor1,y=resid,col=Clutch))+geom_point()+geom_boxplot(aes(group=Factor1),alpha = 0.1)+coord_flip()
Publication methods:
For discrete data and data with non-normal distributions, generalized linear mixed modelling was used (GLMM) (lme4, Douglas et al. 2015; glmmADMB Fournier et al. 2012 & Skaug et al. 2016). Again “Clutch” was treated as a random effect modelled with random intercepts for all models. Appropriate families were selected based on the data distribution. Numerous families and link functions were evaluated where necessary and the optimal parameters were selected based on the Akaike information criterion (AIC). For mobile or non-mobile (yes/no) data a logistic regression was used, for count data (number of cells) a negative binomial family was selected. The significance of inclusion of an independent variable or interaction terms were evaluated using log-likelihood ratio. Holm-Sidak post-hocs were then performed for pair-wise comparisons using the least square means (LSmeans, Russel 2016). Pearson residuals were evaluated graphically using predicted vs level plots. All analyses were performed using R (version 3.5.1, R Core Team, 2018).
Douglas Bates, Martin Maechler, Ben Bolker, Steve Walker (2015). Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software, 67(1), 1-48.<doi:10.18637/jss.v067.i01>.
Russell V. Lenth (2016). Least-Squares Means: The R Package lsmeans. Journal of Statistical Software, 69(1), 1-33.<doi:10.18637/jss.v069.i01>
Fournier DA, Skaug HJ, Ancheta J, Ianelli J, Magnusson A, Maunder M, Nielsen A, Sibert J (2012). “AD Model Builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models.” _Optim. Methods Softw._, *27*, 233-249.
Skaug H, Fournier D, Bolker B, Magnusson A, Nielsen A (2016). _Generalized Linear Mixed Models using ‘AD Model Builder’_. R package version 0.8.3.3.
R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Power Calculator Cohen’s D
A Cohen’s D is a standardized effect size which is defined as the difference between your two groups measured in standard deviations. Because the Cohen’s D unit is standard deviations, it can be used when you have no pilot data. As a general guide a Cohen’s D of 0.3, 0.5 and 0.8 corresponds to mild, moderate and large effect sizes, respectively. However, this depends on the context; for example biomarkers would want the effect size to be at least 2, as biomarkers aren’t useful when the groups overlap.
Example paragraph:
As no pilot data has been collected for these experiments, a standardized effect size was used for the power analysis. For this pilot study we will be aiming to detect a large clinically relevant effect size with a Cohen’s d of 0.8. A power analysis using the two-tailed student’s t-test, Sidak corrected for 3 comparisons, with an alpha of 0.05 and a power of 0.8 was performed. From this analysis it was found that 35 human samples in each group would be required.
Cohen J. 1988. Statistical Power Analysis for the Behavioral Sciences. 2nd ed.Hillsdale: Lawrence Erlbaum Associates.
Power Calculator
The calculator is for Sidak corrected multiple t-tests. It’s useful for when you have pilot data and have estimates for the mean of the control group and the standard deviations. It allows you to estimate the number of animals required to detect a range of percentage changes from the control group.
Example paragraph:
A power analysis was performed on the primary measure of probe trial performance in the Morris water maze using an alpha of 0.05, and a power of 0.8. Previously acquired data was used for the analysis which had a mean of 24.59 and a standard deviation of 11.21. The clinically relevant therapeutic effect size of a 60% improvement in the task was chosen. This analysis found an n of 14 mice per group is required for the Sidak corrected post-hoc analysis with 6 comparisons. Accounting for the possibility of 10% attrition an n of 16 was chosen for these experiments.
Cohen J. 1988. Statistical Power Analysis for the Behavioral Sciences. 2nd ed.Hillsdale: Lawrence Erlbaum Associates.
Talk on social media and science
Here is a presentation I did at the ARUK Early Career Researcher conference on the advantages of having an online presence as a scientist.
More info to come soon!
Download the presentation by clicking here – Don’t be a faceless researcher